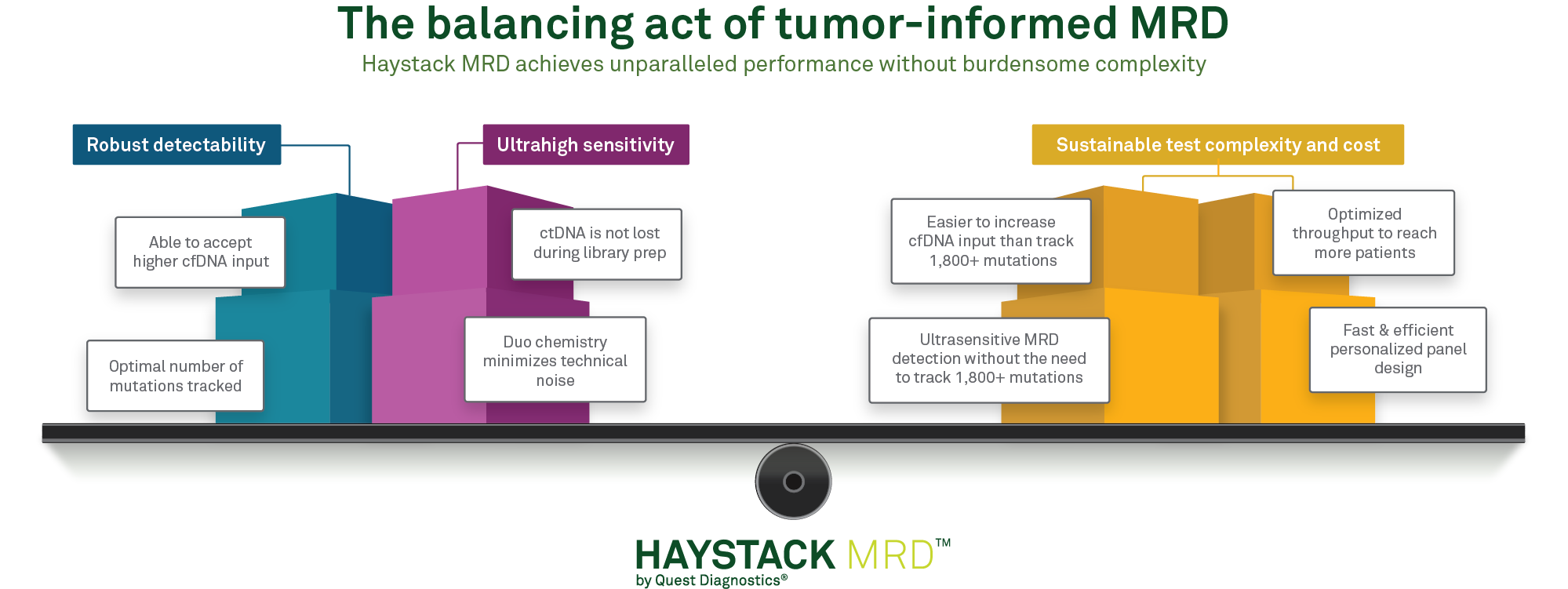
Haystack MRD™ strikes the right balance of DNA molecules interrogated and number of mutations tracked for optimizing test efficiency and sustainability
By increasing sensitivity, minimal residual disease (MRD) tests can confidently detect residual, recurrent, or resistant disease earlier than traditional methods like imaging and antigen testing. For circulating tumor DNA (ctDNA)-based tests such as Haystack MRD, increasing sensitivity-or the ability to detect the smallest quantity of ctDNA in the blood-can be achieved in two ways: by increasing the quantity of ctDNA that is analyzed and/or by increasing the number of mutations tracked.
Higher input
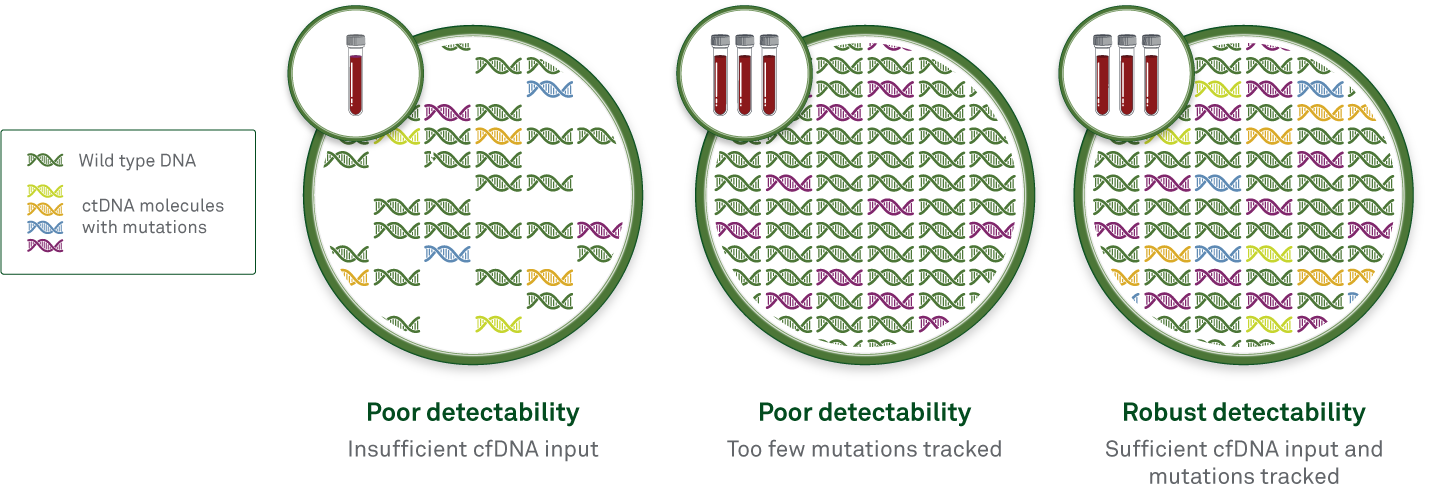
Increased input of ctDNA molecules can be achieved simply by collecting a greater volume of blood from the patient. Some MRD tests are limited to a maximum input of one or two 10-ml blood tubes, whereas Haystack MRD can accept up to 3 tubes per test. Using more input material increases the chances that rare ctDNA molecules will be detected if they are present in the patient’s bloodstream.
Efficient workflow
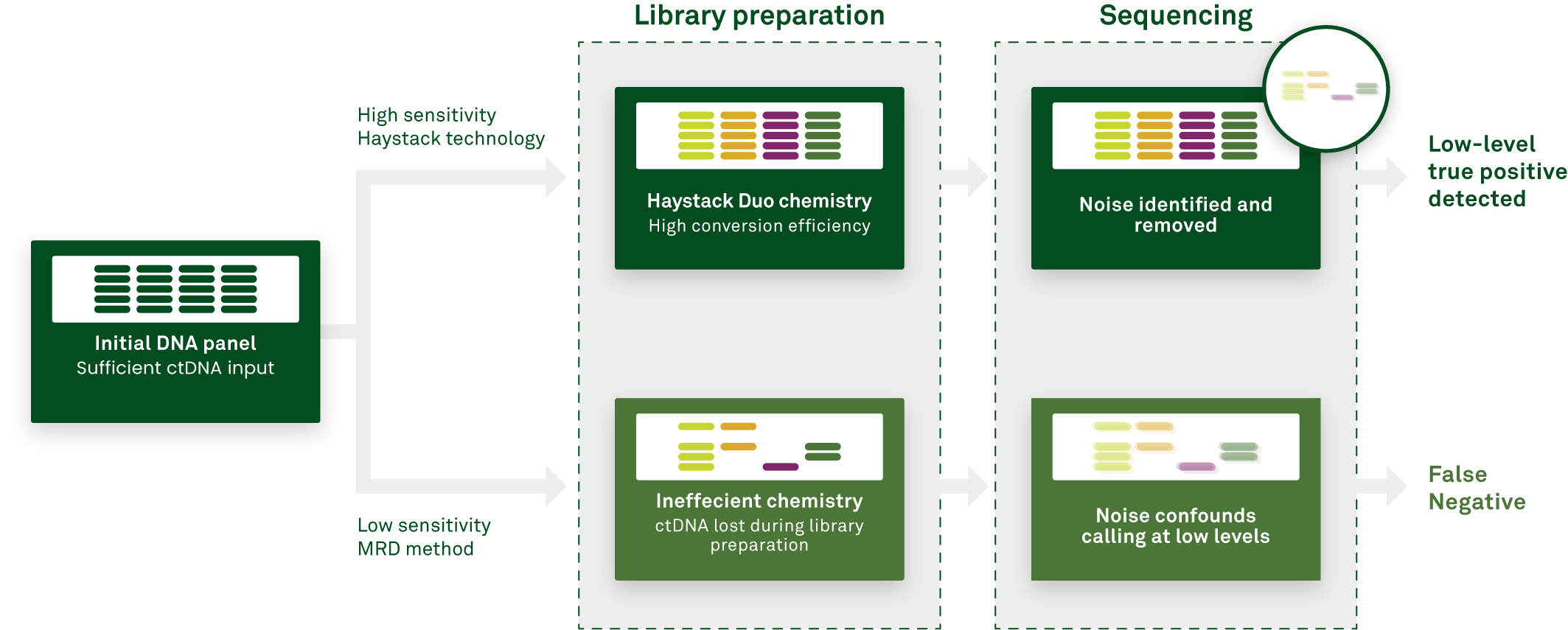
The number of ctDNA molecules may be further increased by improving the efficiency of the assay workflow. ct DNA makes up only a small percentage of the total cell-free DNA (cfDNA) in the bloodstream. After cfDNA is isolated from a blood sample, it is amplified to produce a DNA library that will undergo next-generation sequencing (NGS) to detect mutations that indicate MRD is present.
Haystack’s technology, is designed to preferentially convert ctDNA into library, decreasing the competition with genomic DNA that and increasing the number of ctDNA molecules that are ultimately available for input into the sequencing step of the assay.
Optimal number of mutations tracked
Sensitivity can also be increased by tracking more mutations-to a certain extent. A caveat is that as more mutations are tracked, more technical noise is naturally introduced in the form of sequencing errors. With increased noise, the MRD calling threshold must be set higher, decreasing sensitivity.
Haystack avoids this potential pitfall by keeping noise low, with less than one error for every 1,000,000 cfDNA molecules. This extremely low error rate translates to a low MRD calling threshold, enabling detection of as few as one mutant molecule in a million DNA molecules. Therefore, Haystack MRD can track up to 50 mutations without introducing troublesome background noise.
Why not track more mutations7 Increasing the number of mutations can have drawbacks. As more mutations are tracked, workflow complexity and sequencing cost increase accordingly. Most solid tumors harbor no more than 50 mutations, so tracking 50 mutations balances test cost and efficiency while providing exceptional sensitivity and specificity.
Exceptional sensitivity
The result is an MRD test that has exceptional performance, with good turnaround times and is competitively priced. To learn more about how Haystack MRD’s balanced approach, click here.